FLEUR code plugin
Description
The FleurCalculation runs Fleur executable e.g.
fleur or fleur_MPI.
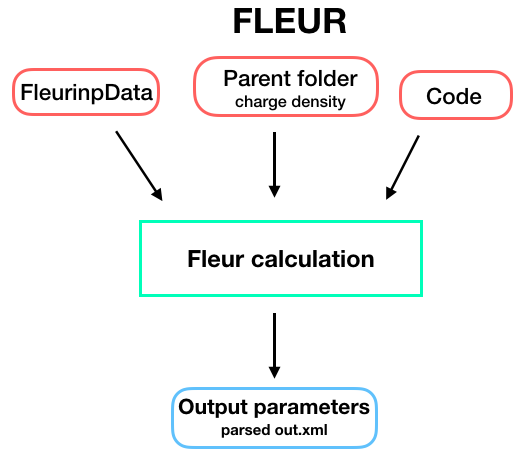
Inputs
To set up an input dictionary, consider using
get_inputs_fleur() which assembles input nodes
in a ready-to-use single dictionary.
The table below shows all possible inputs for the FleurCalculation:
name |
type |
description |
required |
|---|---|---|---|
code |
Code |
Fleur code |
yes |
fleurinp |
FleurinpData |
Object representing inp.xml |
no |
parent_folder |
RemoteData |
Remote folder of another calculation |
no |
settings |
Dict |
special settings |
no |
metadata.options |
Dict |
computational resources |
yes |
fleurinp:
FleurinpData, optional - Data structure which represents the inp.xml file and everything a Fleur calculation needs. For more information see FleurinpData.parent_folder:
RemoteData, optional - If specified, certain files in the previous Fleur calculation folder are copied in the new calculation folder.
Note
fleurinp and parent_folder are both optional. Depending on the setup of the inputs, one of five scenarios will happen:
fleurinp: files belonging to fleurinp will be used as input for FLEUR calculation.
fleurinp + parent_folder (FLEUR): files, given in fleurinp will be used as input for FLEUR calculation. Moreover, initial charge density will be copied from the folder of the parent calculation.
parent_folder (FLEUR): Copies inp.xml file and initial charge density from the folder of the parent FLEUR calculation.
parent_folder (input generator): Copies inp.xml file from the folder of the parent inpgen calculation.
parent_folder (input generator) + fleurinp: files belonging to fleurinp will be used as input for FLEUR calculation. Remote folder is ignored.
Outputs
The table below shows all the output nodes generated by
FleurCalculation:
name |
type |
comment |
|---|---|---|
output_parameters |
Dict |
contains parsed out.xml |
remote_folder |
FolderData |
represents calculation folder |
retrieved |
FolderData |
represents retrieved folder |
All the outputs can be found in calculation.outputs.
remote_folder:
RemoteData- RemoteData which represents the calculation folder on the remote machine.retrieved:
FolderData- FolderData which represents the retrieved folder on the remote machine.output_parameters:
Dict- Contains all kinds of information of the calculation and some physical quantities of the last iteration.An example output node:
# -*- coding: utf-8 -*- (aiidapy)% verdi data dict show 425 { 'CalcJob_uuid': 'a6511a00-7759-484a-839d-c100dafd6118', 'bandgap': 0.0029975592, 'bandgap_units': 'eV', 'charge_den_xc_den_integral': -3105.2785777045, 'charge_density1': 3.55653e-05, 'charge_density2': 6.70788e-05, 'creator_name': 'fleur 27', 'creator_target_architecture': 'GEN', 'creator_target_structure': ' ', 'density_convergence_units': 'me/bohr^3', 'end_date': { 'date': '2019/07/17', 'time': '12:50:27' }, 'energy': -4405621.1469633, 'energy_core_electrons': -99592.985569309, 'energy_hartree': -161903.59225823, 'energy_hartree_units': 'Htr', 'energy_units': 'eV', 'energy_valence_electrons': -158.7015525598, 'fermi_energy': -0.2017877885, 'fermi_energy_units': 'Htr', 'force_largest': 0.0, 'magnetic_moment_units': 'muBohr', 'magnetic_moments': [ 2.7677822875, 2.47601e-05, 2.22588e-05, 6.05518e-05, 0.0001608849, 0.0001504687, 0.0001321699, -3.35528e-05, 1.87169e-05, -0.0002957294 ], 'magnetic_spin_down_charges': [ 5.8532354421, 6.7738647125, 6.8081938915, 6.8073232631, 6.8162583243, 6.8156475799, 6.8188399492, 6.813423175, 6.7733972589, 6.6797683064 ], 'magnetic_spin_up_charges': [ 8.6210177296, 6.7738894726, 6.8082161503, 6.8073838149, 6.8164192092, 6.8157980486, 6.8189721191, 6.8133896222, 6.7734159758, 6.679472577 ], 'number_of_atom_types': 10, 'number_of_atoms': 10, 'number_of_iterations': 49, 'number_of_iterations_total': 49, 'number_of_kpoints': 240, 'number_of_species': 1, 'number_of_spin_components': 2, 'number_of_symmetries': 2, 'orbital_magnetic_moment_units': 'muBohr', 'orbital_magnetic_moments': [], 'orbital_magnetic_spin_down_charges': [], 'orbital_magnetic_spin_up_charges': [], 'output_file_version': '0.27', 'overall_charge_density': 7.25099e-05, 'parser_info': 'AiiDA Fleur Parser v0.2beta', 'parser_warnings': [], 'spin_density': 7.91911e-05, 'start_date': { 'date': '2019/07/17', 'time': '10:38:24' }, 'sum_of_eigenvalues': -99751.687121869, 'title': 'A Fleur input generator calulation with aiida', 'unparsed': [], 'walltime': 7923, 'walltime_units': 'seconds', 'warnings': { 'debug': {}, 'error': {}, 'info': {}, 'warning': {} } }
Errors
Errors of the parsing are reported in the log of the calculation (accessible
with the verdi process report command).
Everything that Fleur writes into stderr is also shown here, i.e all JuDFT error messages.
Example:
(aiidapy)% verdi process report 513
*** 513 [scf: fleur run 1]: None
*** (empty scheduler output file)
*** (empty scheduler errors file)
*** 3 LOG MESSAGES:
+-> ERROR at 2019-07-17 14:57:01.108964+00:00
| parser returned exit code<302>: FLEUR calculation failed.
+-> ERROR at 2019-07-17 14:57:01.097337+00:00
| FLEUR calculation did not finishsuccessfully.
+-> WARNING at 2019-07-17 14:57:01.056220+00:00
| The following was written into std error and piped to out.error :
| I/O warning : failed to load external entity "relax.xml"
| rm: cannot remove ‘cdn_last.hdf’: No such file or directory
| **************juDFT-Error*****************
| Error message:e>vz0
| Error occurred in subroutine:vacuz
| Hint:Vacuum energy parameter too high
| Error from PE:0/24
Moreover, all warnings and errors written by Fleur in the out.xml file are stored in the
ParameterData under the key warnings, and are accessible with Calculation.res.warnings.
More serious FLEUR calculation failures generate a non-zero exit code. Each exit code has it’s own reason:
Exit code |
Reason |
|---|---|
300 |
One of output files can not be opened |
301 |
No retrieved folder found |
302 |
FLEUR calculation failed for unknown reason |
303 |
XML output file was not found |
304 |
Parsing of XML output file failed |
305 |
Parsing of relax XML output file failed |
310 |
FLEUR calculation failed due to memory issue |
311 |
FLEUR calculation failed because atoms spilled to the vacuum |
312 |
FLEUR calculation failed due to MT overlap |
313 |
FLEUR calculation failed due to MT overlap during relaxation |
314 |
Problem with cdn is suspected |
315 |
Invalid Elements found in the LDA+U density matrix. |
316 |
Calculation failed due to time limits. |
Parallelization options
For parallel FLEUR calculations the input under metadata.options can be used.
In higher level workchains this input might be present as a plain options input,
but it is completely equivalent to the metadata.options input.
inputs.metadata.options = {
'resources': {
'num_machines': 2, #Number of computing nodes
'num_mpiprocs_per_machine': 4, #Number of MPI processes per node
'num_cpus_per_mpiproc': 12, #Number of OMP threads per MPI process
},
'withmpi': True, #This flag makes sure that the process is submitted using MPI
'max_wallclock_seconds': 3600, #Maximum wallclock time in seconds
}
This will result in setting the following slurm Parallelization variables in the submit script.
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=6
#SBATCH --cpus-per-task=8
#SBATCH --time=01:00:00
#... Further configuration options unrelated to parallelization...
'srun' '/path/to/fleur/' '<further FLEUR cmdline flags, e.g. -last_extra>'
Note, that the srun command is computer specific and is configured in verdi computer setup
with the Mpirun command option.
Additional advanced features
In general see the FLEUR documentation.
While the input link with name fleurinp is used for the content of the
inp.xml, additional parameters for changing the plugin behavior, can be specified in the
settings input, also of type Dict.
Below we summarise some of the options that you can specify, and their effect.
In each case, after having defined the content of settings_dict, you can use
it as input of a calculation calc by doing:
calc.use_settings(Dict(dict=settings_dict))
Adding command-line options
If you want to add command-line options to the executable (particularly relevant e.g. ‘-hdf’ use hdf, or ‘-magma’ use different libraries, magma in this case), you can pass each option as a string in a list, as follows:
settings_dict = {
'cmdline': ['-hdf', '-magma'],
}
The default command-line of a fleur execution of the plugin looks like this for the torque scheduler:
'mpirun' '-np' 'XX' 'path_to_fleur_executable' '-wtime' 'XXX' < 'inp.xml' > 'shell.out' 2> 'out.error'
If the code node description contains ‘hdf5’ in some form, the plugin will use per default hdf5, it will only copy the last hdf5 density back, not the full cdn.hdf file. The Fleur execution line becomes in this case:
'mpirun' '-np' 'XX' 'path_to_fleur_executable' '-last_extra' '-wtime' 'XXX' < 'inp.xml' > 'shell.out' 2> 'out.error'
Retrieving more files
AiiDA-FLEUR does not copy all output files generated by a FLEUR calculation. By default, the plugin
copies only out.xml, cdn1 and inp.xml and other technical files.
Depending on certain switches in used inp.xml, the plugin
is capable of automatically adding additional files to the copy list:
if
band=T:bands.1,bands.2if
dos=T:DOS.1,DOS.2if
pot8=T:pot*if
l_f=T:relax.xml
If you know that your calculation is producing additional files that you want to
retrieve (and preserve in the AiiDA repository in the long term), you can add
those files as a list as follows (here in the case of a file named
testfile.txt):
settings_dict = {
'additional_retrieve_list': ['testfile.txt'],
}
Retrieving less files
If you know that you do not want to retrieve certain files(and preserve in the AiiDA repository
in the long term). i.e. the cdn1 file is to large and it is stored somewhere else anyway,
you can add those files as a list as follows (here in the case of a file named
testfile.txt):
settings_dict = {
'remove_from_retrieve_list': ['testfile.txt'],
}
Copy more files remotely
The plugin copies by default the mixing_history* files if a parent_folder is given
in the input.
If you know that for your calculation you need some other files on the remote machine, you can add
those files as a list as follows (here in the case of a file named
testfile.txt):
settings_dict = {
'additional_remotecopy_list': ['testfile.txt'],
}
Copy less files remotely
If you know that for your calculation do not need some files which are copied per default by
the plugin you can add those files as a list as follows (here in the case of a file named
testfile.txt):
settings_dict = {
'remove_from_remotecopy_list': ['testfile.txt'],
}