FleurinpData
Class:
FleurinpDataString to pass to the
DataFactory():fleur.fleurinpAim: store input files for the FLEUR code and provide user-friendly editing.
What is stored in the database: the filenames, a parsed inp.xml files as nested dictionary
What is stored in the file repository: inp.xml file and other optional files.
Additional functionality: Provide user-friendly methods. Connected to structure and Kpoints AiiDA data structures
Description/Features
FleurinpData is an additional AiiDA data structure which
represents everything a FleurCalculation
needs, which is mainly a complete inp.xml file.
Note
Currently, FleurinpData methods support
ONLY inp.xml files, which have
everything in them (kpoints, energy parameters, …), i.e which were created with
the -explicit inpgen command line switch.
In general it was designed to account for several separate files too,
but this is no the default way Fleur should be used with AiiDA.
FleurinpData was implemented
to make the plugin more user-friendly, hide complexity and
ensure the connection to AiiDA data structures (StructureData,
KpointsData).
More detailed information about the methods can be found below and in the module code documentation.
Note
If you want to change the input file use the
FleurinpModifier (FleurinpModifier)
class, because a FleurinpData object
has to be stored in the database and usually sealed.
Initialization:
from aiida_fleur.data.fleurinp import FleurinpData
# or FleurinpData = DataFactory('fleur.fleurinp')
F = FleurinpData(files=['path_to_inp.xml_file', <other files>])
# or
F = FleurinpData(files=['inp.xml', <other files>], node=<folder_data_pk>)
If the node attribute is specified, AiiDA will try to get files from the
FolderData corresponding
to the node. If not, it tries to find an inp.xml file using an absolute path
path_to_inp.xml_file.
Be aware that the inp.xml file name has to be named ‘inp.xml’, i.e. no file names are
changed because the filenames will not be changed before submitting a Fleur Calculation.
If you add another inp.xml file the first one will be overwritten.
Properties
inp_dict: Returns the inp_dict (the representation of the inp.xml file) as it will or is stored in the database.
files: Returns a list of files, which were added to FleurinpData. Note that all of these files will be copied to the folder where FLEUR will be run.
inp_version: Returns the version of the storedinp.xml
parser_info: Returns errors, warnings and information encountered while constructing theinp_dictfrom theinp.xml
Note
FleurinpData will use the masci-tools library to parse the inp.xml. This library contains the schema files for the fleur input and output XML files for many of the fleur releases starting from version 0.27. If a version is encountered that is not yet stored in the installed version of the masci-tools library, the latest available version is used.
User Methods
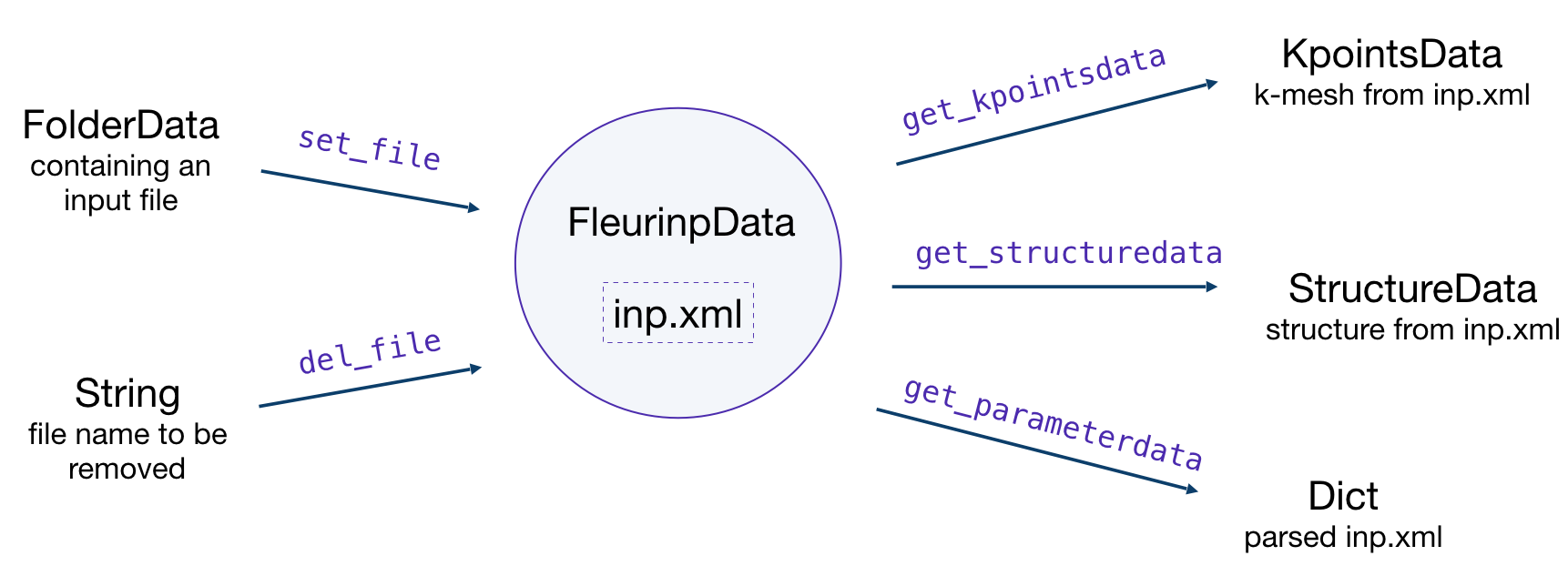
del_file()- Deletes a file fromFleurinpDatainstance.
set_file()- Adds a file from a folder node toFleurinpDatainstance.
set_files()- Adds several files from a folder node toFleurinpDatainstance.
get_fleur_modes()- Analyses the inp.xml and get a dictionary with the corresponding calculation modes (Noco, SOC, …)
get_structuredata()- A CalcFunction which returns an AiiDAStructureDatatype extracted from the inp.xml file.
get_kpointsdata()- A CalcFunction which returns an AiiDAKpointsDatatype produced from the inp.xml. If multiple k-point sets are defined (Fleur release MaX5 or later) a dictionary ofKpointsDatatypes is returned file. This only works if the kpoints are listed in the in inp.xml.
get_parameterdata()- A CalcFunction that extracts aDictnode containing FLAPW parameters. This node can be used as an input for inpgen.
Setting up atom labels
Label is a string that marks a certain atom in the inp.xml file. Labels are created automatically
by the inpgen, however, in some cases it is helpful to control atom labeling. This can be done by
setting up the kind name while initialising the structure:
structure = StructureData(cell=cell)
structure.append_atom(position=(0.0, 0.0, -z), symbols='Fe', name='Fe123')
structure.append_atom(position=(x, y, 0.0), symbols='Pt')
structure.append_atom(position=(0., 0., z), symbols='Pt')
in this case both of the Pr atoms will get default labels, but ‘Fe’ atom will the label ‘123’ (actually ‘ 123’, but all of the methods in AiiDA-Fleur are implemented in a way that user should know only last digit characters).
Note
Kind name, which is used for labeling, must begin from the element name and end up with a number.
It is very important that the first digit of the number is smaller than 4: 152, 3, 21
can be good choices, when 492, 66, 91 are forbidden.
Warning
Except setting up the label, providing a kind name also creates a new specie. This is because
the 123 will not only appear as a label, but also in the atom number. In our case, the line
in the inpgen input corresponding to Fe atom will look like 26.123 0 0 -z 123. Hence,
if we would have another Fe atom with the default kind name, both of the Fe atom would belong
to different atom group, generally resulting in lower symmetry.
Given labels can be used for simplified xml methods. For example,
when one performs SOC calculations it might be needed to vary socscale parameter for a certain
atom. Knowing the correct label of the atom, it is straightforward to make such a change in
FleurinpData object
(using the FleurinpModifier)
or pass a corresponding
line to inpxml_changes of workchain parameters:
# an example of inpxml_changes list, that sets socscale of the iron atom
# from the above structure to zero
inpxml_changes = [('set_species_label', {'at_label': '123',
'attributedict': {
'special': {'socscale': 0.0}
},
'create': True
})]
# in this example the atomgroup, to which the atom with label '222' belongs,
# will be modified
fm = FleurinpModifier(SomeFleurinp)
fm.set_atomgroup_label({'force': {'relaxXYZ': 'FFF'}, atom_label=' 222')