FleurinpModifier
Contents
Description
The FleurinpModifier class has
to be used if you want to change anything in a stored
FleurinpData.
It will store and validate all the changes you wish to do and produce a new
FleurinpData node
after you are done making changes and apply them.
FleurinpModifier provides a
user with methods to change the Fleur input.
In principle a user can do everything, since he could prepare a FLEUR input himself and create a
FleurinpData object from that input.
Note
In the open provenance model nodes stored in the database
cannot be changed (except extras and comments). Therefore, to modify something in a stored
inp.xml file one has to create a new FleurinpData
which is not stored, modify it and store it
again. However, this node would pop into existence unlinked in the database and this would mean
we loose the origin from what data it comes from and what was done to it. This is the task of
FleurinpModifier.
Usage
To modify an existing FleurinpData, a
FleurinpModifier instance
has to be initialised staring from the
FleurinpData instance.
After that, a user should register
certain modifications which will be cached and can be previewed. They will be applied on
a new FleurinpData
object when the freeze method is executed. A code example:
from aiida_fleur.data.fleurinpmodifier import FleurinpModifier
F = FleurinpData(files=['inp.xml'])
fm = FleurinpModifier(F) # Initialise FleurinpModifier class
fm.set_inpchanges({'dos' : True, 'Kmax': 3.9 }) # Add changes
fm.show() # Preview
new_fleurinpdata = fm.freeze() # Apply
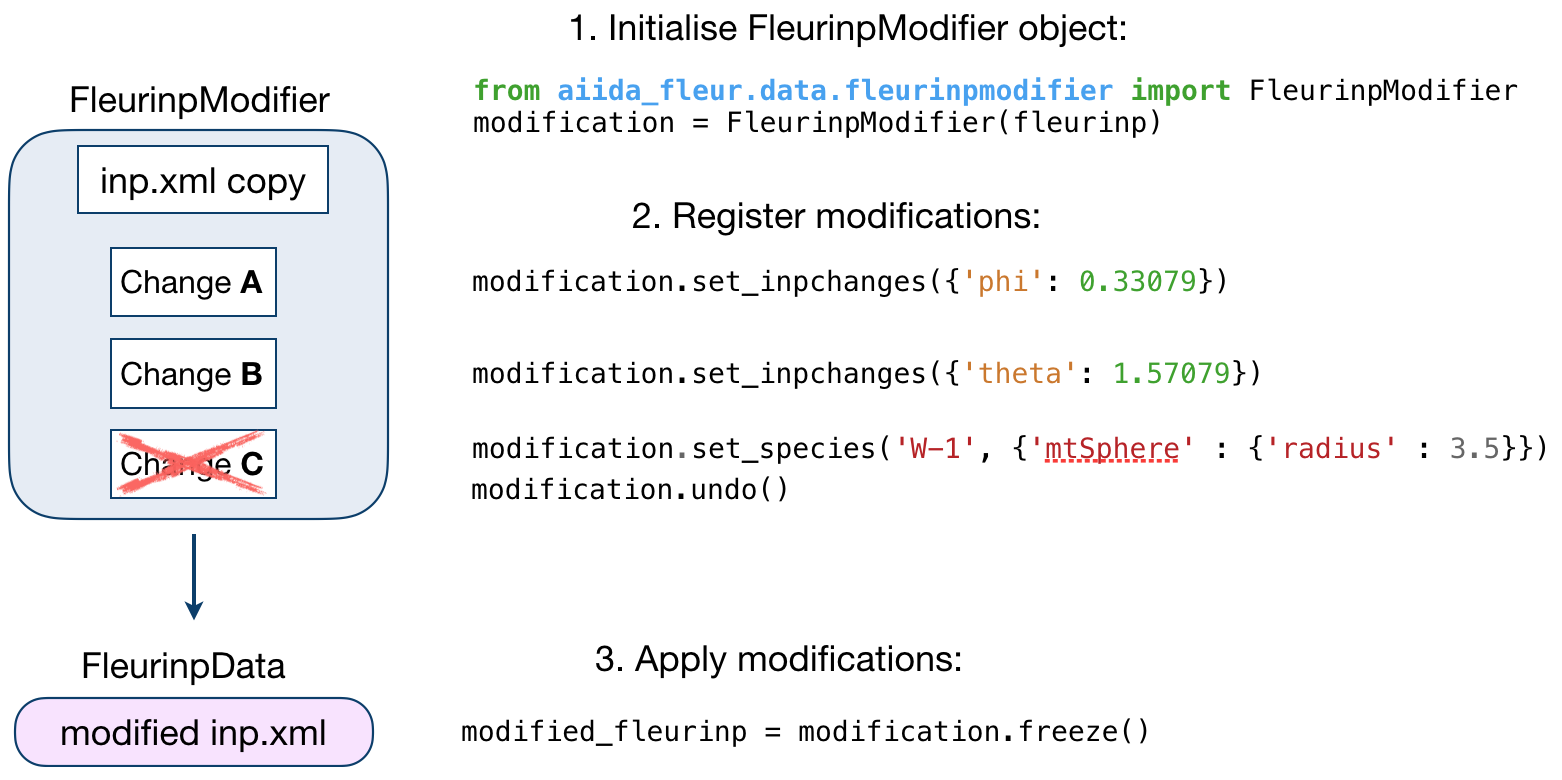
The figure below illustrates the work of the
FleurinpModifier class.
User Methods
General methods
validate(): Tests if the changes in the given list are validated.
freeze(): Applies all the changes in the list, callsmodify_fleurinpdata()and returns a newFleurinpDataobject.
changes(): Displays the current list of changes.
undo(): Remove the last registered change or all registered changes
show(): Applies the modifications and displays/prints the resultinginp.xmlfile. Does not generate a newFleurinpDataobject.
Modification registration methods
The registration methods can be separated into three groups. First of all,
there are XML methods that require deeper knowledge about the structure of an inp.xml file.
All of them require an xpath input:
xml_set_attrib_value_no_create(): Set an attribute on the specified xml elements to the specified value(s). Theoccurrencesargument can be used to select, which occurences to modify
xml_set_text_no_create(): Set the text on the specified xml elements to the specified value(s). Theoccurrencesargument can be used to select, which occurences to modify
xml_create_tag(): Insert an xml element in the xml tree.
xml_replace_tag(): Replace an xml element in the xml tree.
xml_delete_tag(): Delete an xml element in the xml tree.
xml_delete_att(): Delete an attribute on a xml element in the xml tree.
On the other hand, there are shortcut methods that already know some paths:
set_species(): Specific user-friendly method to change species parameters.
set_atomgroup(): Specific method to change atom group parameters.
set_species_label(): Specific user-friendly method to change a specie of an atom with a certain label.
set_atomgroup_label(): Specific method to change atom group parameters of an atom with a certain label.
set_inpchanges(): Specific user-friendly method for easy changes of attribute key value type.
shift_value(): Specific user-friendly method to shift value of an attribute.
shift_value_species_label(): Specific user-friendly method to shift value of an attribute of an atom with a certain label.
set_nkpts(): Specific method to set the number of kpoints. (Only for Max4 and earlier)
set_kpath(): Specific method to set a kpoint path for bandstructures (Only for Max4 and earlier)
set_kpointlist(): Specific method to set the used kpoints via a array of coordinates and weights
set_kpointsdata()- User-friendly method used to writes kpoints of aKpointsDatanode to the inp.xml file. It replaces old kpoints for MaX4 versions and older. for MaX5 and later the kpoints are entered as a new kpoint list
switch_kpointset(): Specific method to switch the used kpoint set. (Only for Max5 and later)
set_attrib_value(): user-friendly method for setting attributes in the xml file by specifying their name
set_first_attrib_value(): user-friendly method for setting the first occurrence of an attribute in the xml file by specifying its name
add_number_to_attrib(): user-friendly method for adding to or multiplying values of attributes in the xml file by specifying their name
add_number_to_first_attrib(): user-friendly method for adding to or multiplying values of the first occurrence of the attribute in the xml file by specifying their name
set_text(): user-friendly method for setting text on xml elements in the xml file by specifying their name
set_first_text(): user-friendly method for setting the text on the first occurrence of an xml element in the xml file by specifying its name
set_simple_tag(): user-friendly method for creating and setting attributes on simple xml elements (only attributes) in the xml file by specifying its name
set_complex_tag(): user-friendly method for creating complex tags in the xml file by specifying its name
create_tag(): User-friendly method for inserting a tag in the right place by specifying it’s name
delete_tag(): User-friendly method for delete a tag by specifying it’s name
delete_att(): User-friendly method for deleting an attribute from a tag by specifying it’s name
replace_tag(): User-friendly method for replacing a tag by another by specifying its name
set_nmmpmat(): Specific method for initializing or modifying the density matrix file for a LDA+U calculation (details see below)
rotate_nmmpmat(): Specific method for rotating a block of the density matrix file for a LDA+U calculation (details see below) in real space
In addition there are methods for manipulating the stored files on the FleurinpData instance directly:
set_file(): Set a file on the Fleurinpdata instance
del_file(): Delete a file on the Fleurinpdata instance
The figure below shows a comparison between the use of XML and shortcut methods.

Warning
Deprecated XML modification methods
After the aiida-fleur release 1.1.4 the FleurinpModifier was restructured to enhance it’s capabilities and to make it more robust. During this process several modification functions were renamed or deprecated. Even though all the old usage is still supported it is encouraged to switch to the new method names and behaviours:
xml_set_attribv_occ(),xml_set_all_attribv()andxml_set_first_attribv()are unified in the methodxml_set_attrib_value_no_create(). However, these functions can no longer create missing subtagsxml_set_text_occ(),xml_set_all_text()andxml_set_text()are unified in the methodxml_set_text_no_create(). However, these functions can no longer create missing subtagscreate_tag()is now a higher-level function. The old function requiring an xpath is now calledxml_create_tag()replace_tag()is now a higher-level function. The old function requiring an xpath is now calledxml_replace_tag()delete_tag()is now a higher-level function. The old function requiring an xpath is now calledxml_delete_tag()delete_att()is now a higher-level function. The old function requiring an xpath is now calledxml_delete_att()an xml element in the xml tree.add_num_to_att()was renamed toadd_number_to_attrib()oradd_number_to_first_attrib(). However, these are also higher-level functions now longer requiring a concrete xpathset_atomgr_att()andset_atomgr_att_label()were renamed toset_atomgroup()andset_atomgroup_label(). These functions now also take the changes in the formattributedict={'nocoParams':{'beta': val}}instead ofattributedict={'nocoParams':[('beta': val)]}
Modifying the density matrix for LDA+U calculations
The above mentioned set_nmmpmat() takes a special
role in the modification registration methods, as the modifications are not done on the inp.xml file but the
density matrix file n_mmp_mat used by Fleur for LDA+U calculations. The resulting density matrix file is stored
next to the inp.xml in the new FleurinpData instance produced by calling
the freeze() method and will be used as the initial
density matrix if a calculation is started from this FleurinpData instance.
The code example below shows how to use this method to add a LDA+U procedure to an atom species and provide an initial guess for the density matrix.
from aiida_fleur.data.fleurinpmodifier import FleurinpModifier
F = FleurinpData(files=['inp.xml'])
fm = FleurinpModifier(F) # Initialise FleurinpModifier class
fm.set_species('Nd-1', {'ldaU': # Add LDA+U procedure
{'l': 3, 'U': 6.76, 'J': 0.76, 'l_amf': 'F'}})
fm.set_nmmpmat('Nd-1', orbital=3, spin=1, occStates=[1,1,1,1,0,0,0]) # Initialize n_mmp_mat file with the states
# m = -3 to m = 0 occupied for spin up
# spin down is initialized with 0 by default
new_fleurinpdata = fm.freeze() # Apply
Note
The n_mmp_mat file is a simple text file with no knowledge of which density matrix block corresponds to which
LDA+U procedure. They are read in the same order as they appear in the inp.xml. For this reason the n_mmp_mat
file can become invalid if one adds/removes a LDA+U procedure to the inp.xml after the n_mmp_mat file was
initialized. To circumvent these problems always remove any existing n_mmp_mat file from the
FleurinpData instance, before adding/removing or modifying the LDA+U configuration.
Furthermore the set_nmmpmat() should always be called
after any modifications to the LDA+U configuration.
Usage in Workflows
The FleurinpModifier class can be used nicely to explicitly
modify the FleurinpData instances in scripts. However, when a inp.xml
file should be modified during the run of a aiida-fleur workflow this class cannot be used directly.
Each workflow specifies a wf_parameters dictionary input (potentially more in sub workflows), which contains
a inpxml_changes entry. This entry can be used to modify the used inputs inside the workchain at points
defined by the workflow itself. The syntax for the inpxml_changes entry is as follows:
Explicit definition
wf_parameters = {
'inpxml_changes': [
('set_inpchanges', {'changes': {'dos': True}}),
('set_species', {'species_name': 'Fe-1', {'changes': {'electronConfig': {'flipspins': True}}}})
]
}
is equivalent to
from aiida_fleur.data.fleurinpmodifier import FleurinpModifier
F = FleurinpData(files=['inp.xml'])
fm = FleurinpModifier(F)
fm.set_inpchanges({'dos': True})
fm.set_species('Fe-1', {'electronConfig': {'flipspins': True}})
Using inpxml_changes()
As can be seen from the above example, the syntax for the inpxml_changes entry is quite verbose,
especially if compared with the more compact formulation using the FleurinpModifier
directly.
For this reason a helper function inpxml_changes() is implemented
to construct the inpxml_changes entry with the exact same syntax as the FleurinpModifier
It is used as a contextmanager, which behaves exactly like the Modifier inside it’s with block and
enters a inpxml_changes entry into the dictionary passed to this function after the with block
is terminated.
from aiida_fleur.data.fleurinpmodifier import inpxml_changes
parameters = {}
with inpxml_changes(parameters) as fm:
fm.set_inpchanges({'dos': True})
fm.set_species('Fe-1', {'electronConfig': {'flipspins': True}})
print(parameters)
from aiida_fleur.data.fleurinpmodifier import inpxml_changes
from aiida import plugins
FleurBandDOS = plugins.WorkflowFactory('fleur.banddos')
inputs = FleurBandDOS.get_builder()
with inpxml_changes(inputs) as fm:
fm.set_inpchanges({'dos': True})
fm.set_species('Fe-1', {'electronConfig': {'flipspins': True}})