Fleur Dzyaloshinskii–Moriya Interaction energy workchain
Current version: 0.2.0
Class:
FleurDMIWorkChainString to pass to the
WorkflowFactory():fleur.dmiWorkflow type: Scientific workchain, force theorem sub-group
Contents
Import Example:
from aiida_fleur.workflows.dmi import FleurDMIWorkChain
#or
WorkflowFactory('fleur.dmi')
Description/Purpose
This workchain calculates Dzyaloshinskii–Moriya Interaction energy over a given set of q-points.
In this workchain the force-theorem is employed which means the workchain converges
a reference charge density first
then it submits a single FleurCalculation with a <forceTheorem> tag. However, it is possible
to specify inputs to use external pre-converged charge density and use is as a reference.
The task of the workchain us to calculate the energy difference between two or several structures having a different magnetisation profile (theta and phi values can also vary):
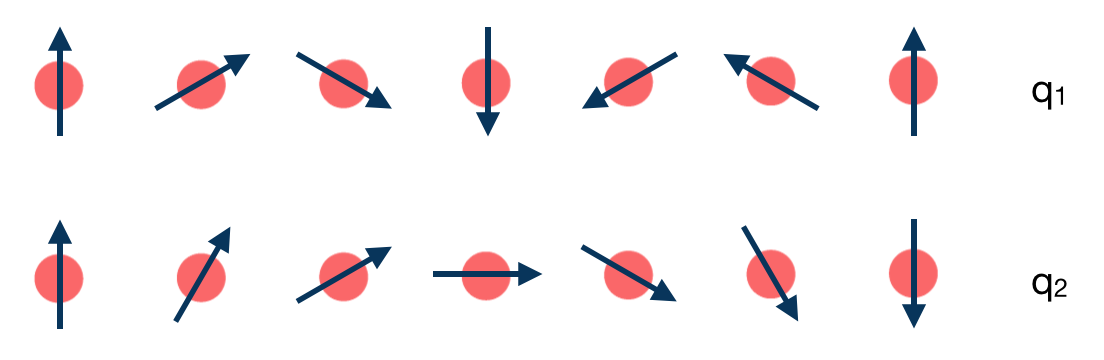
To do this, the workchain employs the force theorem approach:
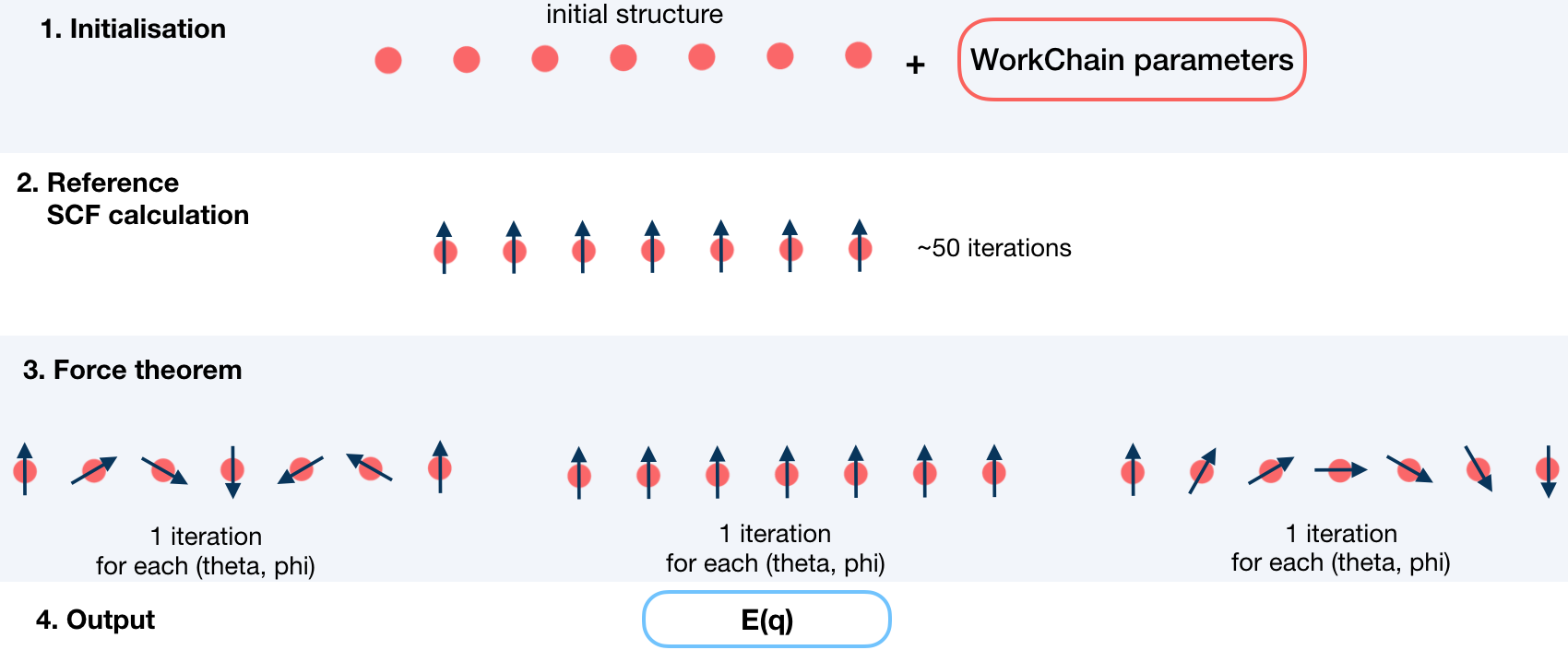
It is not always necessary to start with a structure. Setting up input parameters correctly (see Supported input configurations) one can start from a given FleuinpData, inp.xml or converged/not-fully-converged reference charge density.
Input nodes
The FleurSSDispWorkChain employs
exposed feature of the AiiDA, thus inputs for the nested
SCF workchain should be passed in the namespace
scf.
name |
type |
description |
required |
|---|---|---|---|
scf |
namespace |
inputs for nested SCF WorkChain |
no |
fleur |
Fleur code |
yes |
|
wf_parameters |
Settings of the workchain |
no |
|
fleurinp |
no |
||
remote |
Remote folder of another calculation |
no |
|
options |
AiiDA options (computational resources) |
no |
Only fleur input is required. However, it does not mean that it is enough to specify fleur only. One must keep one of the supported input configurations described in the Supported input configurations section.
Workchain parameters and its defaults
wf_parameters
wf_parameters: Dict - Settings of the workflow behavior. All possible
keys and their defaults are listed below:
# -*- coding: utf-8 -*-
'beta': {'all': 1.57079}, # see description below
'sqas_theta': [0.0, 1.57079, 1.57079], # a list of theta values for the FT
'sqas_phi': [0.0, 0.0, 1.57079], # a list of phi values for the FT
'soc_off': [], # a list of atom labels to switch off SOC term
'q_vectors': [[0.0, 0.0, 0.0], # set a set of q-vectors to calculate DMI dispersion
[0.1, 0.1, 0.0]]
'add_comp_para': {
'only_even_MPI': False, # True if suppress parallelisation having odd number of MPI
'max_queue_nodes': 20, # Max number of nodes allowed (used by automatic error fix)
'max_queue_wallclock_sec': 86400 # Max number of walltime allowed (used by automatic error fix)
},
'ref_qss': [0.0, 0.0, 0.0], # sets a q-vector for the reference calculation
'inpxml_changes': [], # additional changes before the FT step
beta is a python dictionary containing a key: value pairs. Each pair sets beta parameter
in an inp.xml file. key specifies the atom label to change, key equal to ‘all’ sets all
atoms groups. For example,
'beta' : {'222' : 1.57079}
changes
<atomGroup species="Fe-1">
<filmPos label=" 222">.0000000000 .0000000000 -11.4075100502</filmPos>
<force calculate="T" relaxXYZ="TTT"/>
<nocoParams l_relax="F" alpha=".00000000" beta="0.00000" b_cons_x=".00000000" b_cons_y=".00000000"/>
</atomGroup>
to:
<atomGroup species="Fe-1">
<filmPos label=" 222">.0000000000 .0000000000 -11.4075100502</filmPos>
<force calculate="T" relaxXYZ="TTT"/>
<nocoParams l_relax="F" alpha=".00000000" beta="1.57079" b_cons_x=".00000000" b_cons_y=".00000000"/>
</atomGroup>
Note
beta actually sets a beta parameter for a whole atomGroup. It can be that the atomGroup correspond to several atoms and beta switches sets beta for atoms that was not intended to change. You must be careful and make sure that several atoms do not correspond to a given specie.
soc_off is a python list containing atoms labels. SOC is switched off for species, corresponding to the atom with a given label.
Note
It can be that the spice correspond to several atoms and soc_off switches off SOC for atoms that was not intended to change. You must be careful and make sure that several atoms do not correspond to a given specie.
An example of soc_off work:
'soc_off': ['458']
changes
<species name="Ir-2" element="Ir" atomicNumber="77" coreStates="17" magMom=".00000000" flipSpin="T">
<mtSphere radius="2.52000000" gridPoints="747" logIncrement=".01800000"/>
<atomicCutoffs lmax="8" lnonsphr="6"/>
<energyParameters s="6" p="6" d="5" f="5"/>
<prodBasis lcutm="4" lcutwf="8" select="4 0 4 2"/>
<lo type="SCLO" l="1" n="5" eDeriv="0"/>
</species>
-----
<atomGroup species="Ir-2">
<filmPos label=" 458">1.000/4.000 1.000/2.000 11.4074000502</filmPos>
<force calculate="T" relaxXYZ="TTT"/>
<nocoParams l_relax="F" alpha=".00000000" beta=".00000000" b_cons_x=".00000000" b_cons_y=".00000000"/>
</atomGroup>
to:
<species name="Ir-2" element="Ir" atomicNumber="77" coreStates="17" magMom=".00000000" flipSpin="T">
<mtSphere radius="2.52000000" gridPoints="747" logIncrement=".01800000"/>
<atomicCutoffs lmax="8" lnonsphr="6"/>
<energyParameters s="6" p="6" d="5" f="5"/>
<prodBasis lcutm="4" lcutwf="8" select="4 0 4 2"/>
<special socscale="0.0"/>
<lo type="SCLO" l="1" n="5" eDeriv="0"/>
</species>
As you can see, I was careful about “Ir-2” specie and it contained a single atom with a label 458. Please also refer to Setting up atom labels section to learn how to set labels up.
sqas_theta and sqas_phi are python lists that set SOC theta and phi values.
prop_dir is used only to set up a spin spiral propagation direction to
calc_parameters['qss'] which will be passed to SCF workchain and inpgen. It can be used
to properly set up symmetry operations in the reference calculation.
options
options: Dict - AiiDA options (computational resources).
Example:
'resources': {"num_machines": 1, "num_mpiprocs_per_machine": 1},
'max_wallclock_seconds': 6*60*60,
'queue_name': '',
'custom_scheduler_commands': '',
'import_sys_environment': False,
'environment_variables': {}
Output nodes
out:Dict- Information of workflow results like success, last result node, list with convergence behavior"angles": 3, "energies": [ 0.0 ], "energy_units": "eV", "errors": [], "info": [], "initial_structure": "35e5058d-161c-4cf9-801e-4eca99e7d7be", "phi": [ 3.1415927, ], "q_vectors": [ [ 0.0, 0.0, 0.0 ], ], "theta": [ 0.0, ], "warnings": [], "workflow_name": "FleurDMIWorkChain", "workflow_version": "0.1.0"Resulting DMI energies are sorted according to theirs q-vector, theta and phi values i.e.
energies[N]corresponds toq_vectors[N],phi[N]andtheta[N].
Supported input configurations
DMI workchain has several input combinations that implicitly define the workchain layout. Only scf, fleurinp and remote nodes control the behaviour, other input nodes are truly optional. Depending on the setup of the inputs, one of several supported scenarios will happen:
scf:
SCF workchain will be submitted to converge the reference charge density which will be followed be the force theorem calculation. Depending on the inputs given in the SCF namespace, SCF will start from the structure or FleurinpData or will continue converging from the given remote_data (see details in SCF WorkChain).
remote:
Files which belong to the remote will be used for the direct submission of the force theorem calculation.
inp.xmlfile will be converted to FleurinpData and charge density will be used as a reference charge density.remote + fleurinp:
Charge density which belongs to remote will be used as a reference charge density, however
inp.xmlfrom the remote will be ignored. Instead, the given fleurinp will be used. The aforementioned input files will be used for direct submission of the force theorem calculation.
Other combinations of the input nodes scf, fleurinp and remote are forbidden.
Warning
One must follow one of the supported input configurations. To protect a user from the workchain misbehaviour, an error will be thrown if one specifies e.g. both scf and remote inputs because in this case the intention of the user is not clear either he/she wants to converge a new charge density or use the given one.
Error handling
A list of implemented exit codes:
Code |
Meaning |
|---|---|
230 |
Invalid workchain parameters |
231 |
Invalid input configuration |
233 |
Input codes do not correspond to fleur or inpgen codes respectively. |
235 |
Input file modification failed. |
236 |
Input file was corrupted after modifications |
334 |
Reference calculation failed. |
335 |
Found no reference calculation remote repository. |
336 |
Force theorem calculation failed. |
Example usage
# -*- coding: utf-8 -*- from aiida.orm import load_node, Dict from aiida.engine import submit from aiida_fleur.workflows.dmi import FleurDMIWorkChain structure = load_node(STRUCTURE_PK) fleur_code = load_node(FLEUR_PK) inpgen_code = load_node(INPGEN_PK) wf_para = Dict(dict={'add_comp_para': { 'only_even_MPI': False, 'max_queue_nodes': 20, 'max_queue_wallclock_sec': 86400 }, 'beta': {'all': 1.57079}, 'sqas_theta': [0.0, 1.57079, 1.57079], 'sqas_phi': [0.0, 0.0, 1.57079], 'soc_off': [], 'q_vectors': [[0.0, 0.0, 0.0], [0.1, 0.1, 0.0]], 'ref_qss': [0.0, 0.0, 0.0], 'inpxml_changes': [] }) options = Dict(dict={'resources': {'num_machines': 1, 'num_mpiprocs_per_machine': 24}, 'queue_name': 'devel', 'custom_scheduler_commands': '', 'max_wallclock_seconds': 60*60}) parameters = Dict(dict={'atom': {'element': 'Pt', 'lmax': 8 }, 'atom2': {'element': 'Fe', 'lmax': 8, }, 'comp': {'kmax': 3.8, }, 'kpt': {'div1': 20, 'div2': 24, 'div3': 1 }}) wf_para_scf = {'fleur_runmax': 2, 'itmax_per_run': 120, 'density_converged': 0.2, 'mode': 'density' } wf_para_scf = Dict(dict=wf_para_scf) options_scf = Dict(dict={'resources': {'num_machines': 2, 'num_mpiprocs_per_machine': 24}, 'queue_name': 'devel', 'custom_scheduler_commands': '', 'max_wallclock_seconds': 60*60}) inputs = {'scf': {'wf_parameters': wf_para_scf, 'structure': structure, 'calc_parameters': parameters, 'options': options_scf, 'inpgen': inpgen_code, 'fleur': fleur_code }, 'wf_parameters': wf_para, 'fleur': fleur_code, 'options': options } res = submit(FleurDMIWorkChain, **inputs)