Fleur Create Magnetic Film workchain
Current version: 0.2.0
Class: FleurCreateMagneticWorkChain
String to pass to the
WorkflowFactory():fleur.create_magneticWorkflow type: Scientific workchain
Contents
Import Example:
from aiida_fleur.workflows.create_magnetic_film import FleurCreateMagneticWorkChain
#or
WorkflowFactory('fleur.create_magnetic')
Description/Purpose
The workchain constructs a relaxed film structure which is ready-to-use in following magnetic workchains: DMI, MAE or SSDisp.
The inputs include information about the substrate (structure type, miller indices of surfaces or vectors forming the primitive unit cell, chemical elements) and deposited material. The main logic of the workchain is depicted on the figure below:

First, the workchain uses EOS workchain to find the equilibrium lattice parameters for the substrate. For now only bcc and fcc lattices are supported. Note, the algorithm always uses conventional unit cells e.g. one gets 4 atoms in the unit cell for fcc lattice (see the figure above).
After the EOS step the workchain constructs a film which will be used for interlayer distance relaxation via the relaxation workchain. The algorithm creates a film using given miller indices and the ground state lattice constant and replaces some layers with another elements given in the input. For now only single-element layer replacements are possible i.e. each resulting layer can be made of a single element. It is not possible to create e.g. B-N monolayer using this workchain.
Finally, using the result of the relaxation workchain, a magnetic structure having no z-reflection symmetry is
constructed. For this the workchain takes first num_relaxed_layers layers from the relaxed structure and attaches
so many substrate layers so there are total_number_layers layers. The final structure is z-centralised.
Input nodes
The FleurCreateMagneticWorkChain employs
exposed feature of the AiiDA-core, thus inputs for the
EOS and relaxation workchains should be passed in the namespaces
eos and relax correspondingly (see example of usage).
Please note that the structure input node
is excluded from the EOS namespace and from the Relax SCF namespace
since corresponding input structures are created within the CreateMagnetic workchain.
name |
type |
description |
required |
|---|---|---|---|
eos |
namespace |
inputs for nested EOS WC. structure input is excluded. |
no |
relax |
namespace |
inputs for nested Relax WC. structure input of SCF sub-namespace is excluded |
no |
wf_parameters |
Settings of the workchain |
no |
|
eos_output |
EOS output dictionary |
no |
|
optimized_structure |
relaxed film structure |
no |
|
distance_suggestion |
interatomic distance suggestion, output of
the |
no |
Similarly to other workchains, FleurCreateMagneticWorkChain behaves differently depending on the input nodes setup. The list of supported input configurations is given in the section Supported input configurations.
Workchain parameters and its defaults
wf_parameters
wf_parameters: Dict - Settings of the workflow behavior. All possible
keys and their defaults are listed below:
# -*- coding: utf-8 -*-
'lattice': 'fcc', # type of the substrate lattice: 'bcc' or 'fcc'
'miller': None, # miller indices to set up planes forming the p.u.c.
'directions': None, # miller indices to set up vectors forming the p.u.c.
'host_symbol': 'Pt', # chemical element of the substrate
'latticeconstant': 4.0, # initial guess for the substrate lattice constant
'size': (1, 1, 5), # sets the size of the film unit cell for relax step
'replacements': {1: 'Fe', # sets the layer number to be replaced by another element
-1: 'Fe'}, # NOTE: negative number means that replacement will take place
# in the last layer
'decimals': 10, # set the accuracy of writing atom positions
'z_coordinate_window': 2, # set how z-coordinates will be rounded before grouping in layers
'pop_last_layers': 1, # number of bottom layers to be removed before relaxation
'hold_layers': None, # a list of layer numbers to be held during the relaxation
'AFM_name': 'FM', # a name of hardcoded AFM structure: FM, AFM_x, AFM_y, AFM_xy
'magnetic_layers': 1, # the total number of magnetic layers (for symmetric films devide by 2)
'AFM_layer_positions': None, # a suggestion of initial layer z coordinates, used if AFM structures are known
# (relaxXYZ = 'FFF')
'last_layer_factor': 0.85, # factor by which interlayer distance between two last layers
# will be multiplied
'first_layer_factor': 0.0, # factor by which interlayer distance between two first layers
# will be multiplied
'total_number_layers': 4, # use this total number of layers
'num_relaxed_layers': 2, # use this number of relaxed interlayer distances
Some of the parameters, which can be set in the workchain parameter dictionary, control how the structure will be created for the relaxation step. The following procedure is used to construct a film for relaxation:
Create a slab using ASE methods. For this following parameters are used:
lattice,millerordirections,host_symbol,sizeandlatticeconstant(or lattice constant fromdistance_suggestioninput node). For more details refer to ase documentation.Remove pop_last_layers last layers. This step can help one to ensure symmetrical film.
Note
z-reflection or inversion symmetries are not ensured by the workchain even if you specify symmetric replacements. Sometimes you need to remove a few layers before replacements. For example, consider the case of fcc (110) film: if
sizeis equal to (1, 1, 4) there are will be 8 layers in the slab since there are 2 layers in the unit cell. That means the x,y positions of the atom in the first layer are equal to (0.0, 0.0) and the 8th layer coordinates are equal to (0.5, 0.5). Thus, to achieve z-reflection symmetry one needs to remove the 8th layer by specifying'pop_last_layers' : 1in the wf parameters.
Replace atom layers according to
replacementsdictionary. The dictionary should consist of INT: STRING pairs, where INT defines the layer number to be replaced (counting from the lowest layers, INT=1 for the first layer and INT=-1 for the last) and STRING defines the element name.Adjust interlayer distances using
distance_suggestion,first_layer_factorandlast_layer_factor. if the input structure has z-reflection symmetry, thenfirst_layer_factoris ignored and thelast_layer_factorcontrols both surface layers. IfAFM_layer_positionsis given and AFM structures are known for the input lattice and directions, then the adjusting procedure will not do it automatically, but simply enforce z-coordinates fromAFM_layer_positions. Read more in the section Structures with known AFM structures.Warning
Adjusting of interlayer distances for non-symmetric films work well only if substrate is positioned above magnetic elements (z-coordinate of substrate atoms are higher than magnetic ones). This can be achieved by using
replacements: {1: 'Fe'}instead ofreplacements: {-1: 'Fe'}.
Mark fixed layers according to
hold_layers.hold_layersis a list of layer number to be marked as fixed during the relaxation step. Similarly to replacements, the 1st layer corresponds to number 1 and the last to -1.
After the structure is relaxed, the final magnetic non-symmetrical structure is constructed. For this
total_number_layers and num_relaxed_layers setting the total number of layers of the number of layers extracted
from the relaxed structure respectively.
Output nodes
magnetic_structure:StructureData- the relaxed film structure.
Supported input configurations
CreateMagnetic workchain has several input combinations that implicitly define the workchain layout. eos, relax, optimized_structure and eos_output are analysed. Depending on the given setup of the inputs, one of four supported scenarios will happen:
eos + relax + distance_suggestion:
The EOS will be used to calculate the equilibrium structure of the substrate, then Relax WC will be used to relax the interlayer distances. Finally, the non-symmetrical magnetic structure will be created. A good choice if there is nothing to begin with. distance_suggestion will be used to guess a better starting interlayer distances before submitting Relax WC.
eos_output + relax + distance_suggestion:
The equilibrium substrate structure will be extracted from the eos_output, then Relax WC will be used to relax the interlayer distances. Finally, the non-symmetrical magnetic structure will be created. A good choice if EOS was previously done for the substrate. distance_suggestion will be used to guess a better starting interlayer distances before submitting Relax WC.
optimized_structure:
optimized_structure will be treated as a result of Relax WC and directly used to construct the final non-symmetrical magnetic structure. A good choice if everything was done except the very last step.
relax:
Relax WC will be submitted using inputs of the namespace, which means one can for instance continue a relaxation procedure. After Relax WC is finished, the non-symmetrical magnetic structure will be created. A good choice if something wrong happened in one of the relaxation steps of another CreateMagnetic workchain submission.
All the other input configuration will end up with an exit code 231, protecting user from misunderstanding.
Error handling
A list of implemented exit codes:
Code |
Meaning |
|---|---|
230 |
Invalid workchain parameters |
231 |
Invalid input configuration |
380 |
Specified substrate is not bcc or fcc, only them are supported |
382 |
Relaxation calculation failed. |
383 |
EOS WorkChain failed. |
Structures with known AFM structures
Warning
The workchain uses define_AFM_structures method to mark spin-up and spin-down atoms. It does not actually set
initial moment, a user is responsible for adding inpxml_changes changing the initial spin. For instance,
for MaX4 FLEUR one can use following inpxml_changes to set initial moments for AFM structure:
'inpxml_changes': [('set_species_label', {'at_label': '49990',
'attributedict': {'magMom': 4.0},
'create': True}),
('set_species_label', {'at_label': '49991',
'attributedict': {'magMom': -4.0},
'create': True})]}
Spin-up atoms are marked by label '49990' and spin-down atoms are marked by '49991'. Please make sure that
these labels are not overwritten, for example by fixing atoms label.
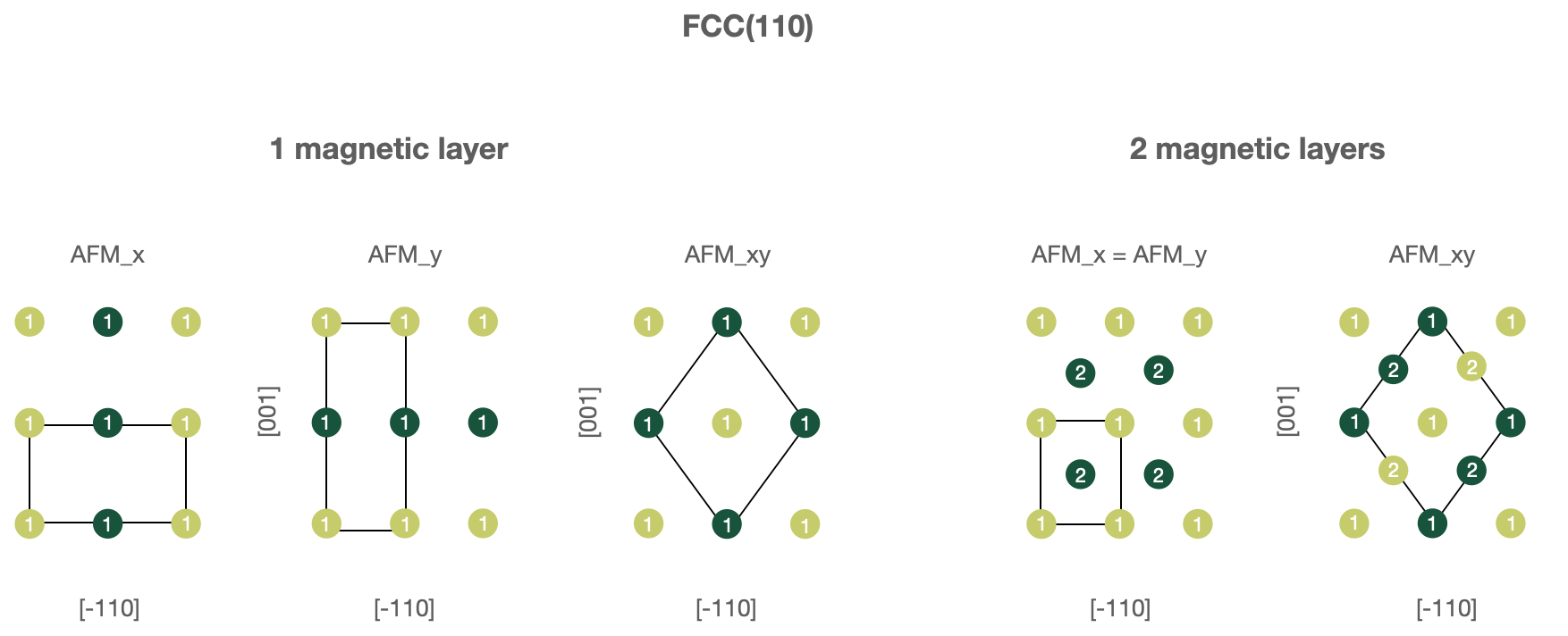
FCC(110) surfaces, 1 or 2 magnetic layers.
In this case one must use following parameters in the
wf_parameters:'lattice': 'fcc' 'directions': [[-1, 1, 0], [0, 0, 1], [1, 1, 0]] 'magnetic_layers': 1 or 2 'AFM_name': 'FM', 'AFM_x', 'AFM_y' or 'AFM_xy'
Note that for
'AFM_xy'structure unit vectors forming a computational cell are changed to[[-1, 1, 2], [1, -1, 2], [1, 1, 0]].
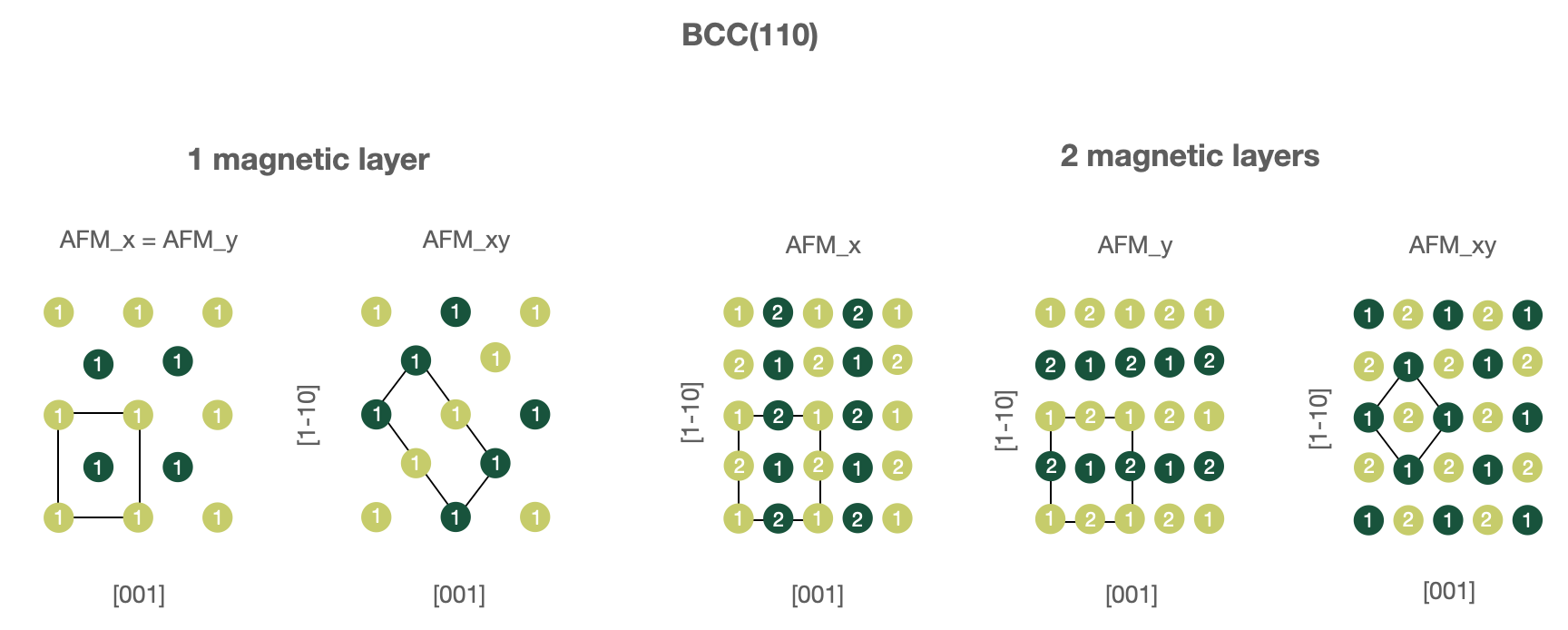
BCC(110) surfaces, 1 or 2 magnetic layers.
In this case one must use following parameters in the
wf_parameters:'lattice': 'bcc' 'directions': [[1, -1, 1], [1, -1, -1], [1, 1, 0]] 'magnetic_layers': 1 or 2 'AFM_name': 'FM', 'AFM_x', 'AFM_y' or 'AFM_xy'
Note that for
'AFM_x'and'AFM_y'structure unit vectors forming a computational cell are changed to[[0, 0, 1], [1, -1, 0], [1, 1, 0]].Warning
The check of input lattice vector directions is hardcoded, which means that the code will not recognize
'bcc'and'[[-1, 1, -1], [-1, 1, 1], [1, 1, 0]]'despite it produces the same structure. In this case the workchain will be excepted.
There is a possibility to enforce layer z-coordinates for generating AFM structures. This allows one to reuse the
relaxed FM structured for following AFM calculation to save computational resources because it is expected that the FM
relaxed structure is a better initial guess for an AFM one rather than a structure, proposed by automatic adjusting
function. To do this, one should make sure that the length of AFM_layer_positions is the same as the total number
of layers of the AFM structure and magnetic_layers is correctly initialised.
Figures below illustrate known AFM structures for FCC(110) and BCC(110) structures. The number on each atoms shows to which layer atom belongs (first or second) and the color corresponds to spin orientation (up or down). Note that the input computational unit cell will be changed (shown on the figures), hence you might want to adjust the k-mesh correspondingly.
Example usage
# -*- coding: utf-8 -*- from aiida.orm import load_node, Dict from aiida.engine import submit from aiida_fleur.workflows.create_magnetic_film import FleurCreateMagneticWorkChain fleur_code = load_node(FLEUR_PK) inpgen_code = load_node(INPGEN_PK) wf_para = { 'lattice': 'fcc', 'directions': [[-1, 1, 0], [0, 0, 1], [1, 1, 0]], 'host_symbol': 'Pt', 'latticeconstant': 4.0, 'size': (1, 1, 5), 'replacements': {0: 'Fe', -1: 'Fe'}, 'decimals': 10, 'pop_last_layers': 1, 'total_number_layers': 8, 'num_relaxed_layers': 3, } wf_para = Dict(dict=wf_para) wf_eos = {'points': 15, 'step': 0.015, 'guess': 1.00 } wf_eos_scf = {'fleur_runmax': 4, 'density_converged': 0.0002, 'itmax_per_run': 50, 'inpxml_changes': [] } wf_eos_scf = Dict(dict=wf_eos_scf) wf_eos = Dict(dict=wf_eos) calc_eos = {'comp': {'kmax': 3.8, }, 'kpt': {'div1': 4, 'div2': 4, 'div3': 4 } } calc_eos = Dict(dict=calc_eos) options_eos = {'resources': {'num_machines': 1, 'num_mpiprocs_per_machine': 4, 'num_cores_per_mpiproc': 6}, 'queue_name': 'devel', 'custom_scheduler_commands': '', 'max_wallclock_seconds': 1*60*60} options_eos = Dict(dict=options_eos) wf_relax = {'film_distance_relaxation': False, 'force_criterion': 0.049, 'relax_iter': 5 } wf_relax_scf = {'fleur_runmax': 5, 'use_relax_xml': True, 'itmax_per_run': 50, 'alpha_mix': 0.015, 'relax_iter': 25, 'force_converged': 0.001, 'force_dict': {'qfix': 2, 'forcealpha': 0.75, 'forcemix': 'straight'}, 'inpxml_changes': [] } wf_relax = Dict(dict=wf_relax) wf_relax_scf = Dict(dict=wf_relax_scf) calc_relax = {'comp': {'kmax': 4.0, }, 'kpt': {'div1': 24, 'div2': 20, 'div3': 1 }, 'atom': {'element': 'Pt', 'rmt': 2.2, 'lmax': 10, 'lnonsph': 6, 'econfig': '[Kr] 5s2 4d10 4f14 5p6| 5d9 6s1', }, 'atom2': {'element': 'Fe', 'rmt': 2.1, 'lmax': 10, 'lnonsph': 6, 'econfig': '[Ne] 3s2 3p6| 3d6 4s2', }, } calc_relax = Dict(dict=calc_relax) options_relax = {'resources': {'num_machines': 1, 'num_mpiprocs_per_machine': 4, 'num_cores_per_mpiproc': 6}, 'queue_name': 'devel', 'custom_scheduler_commands': '', 'max_wallclock_seconds': 1*60*60} inputs = { 'eos': { 'scf': { 'wf_parameters': wf_eos_scf, 'calc_parameters': calc_eos, 'options': options_eos, 'inpgen': inpgen_code, 'fleur': fleur_code }, 'wf_parameters': wf_eos }, 'relax': { 'scf': { 'wf_parameters': wf_relax_scf, 'calc_parameters': calc_relax, 'options': options_relax, 'inpgen': inpgen_code, 'fleur': fleur_code }, 'wf_parameters': wf_relax, }, 'wf_parameters': wf_para } res = submit(FleurCreateMagneticWorkChain, **inputs)